Evolutionary Arms Race Lab
BACKGROUND
If the product of natural selection is adapted organisms, then, over long stretches of time, shouldn’t we expect organisms to approach a state of perfect harmony with their environment? One wrench in the works for this utopian vision is that environments change. This means that yesterday’s adaptation may be today’s mismatch. Moreover, the very process of evolutionary adaptation can lead to environmental change. This can be the case when the "environment" of one species is another species.
Of particular interest are cases where species interact antagonistically. Examples include prey and their predators, hosts and their parasites, as well as plants and their herbivores. In such victim-exploiter relations, the victim should be selected to avoid the exploiter (camouflage, resistance, distastefulness, etc.). However, an evolutionary innovation by a victim sets the stage for a counter-adaptation by the exploiter, so that it can utilize the new victim class. This drives new evolutionary developments in the victim and the cycle continues. This back-and-forth game is termed an evolutionary arms race.
In this lab, you will explore the potential of an evolutionary arms race between a bacterium (the victim) and a virus that infects it (the exploiter). This virus is called a "bacteriophage" or simply "phage" (from the Greek verb phagein meaning "to eat"). You will select for bacteria that are resistant to phage attack. Then you will select for phage that are able to infect these resistant bacteria. You will also determine whether there are costs involved in proceeding down an evolutionary arms race. For instance, does a bacterium pay a cost to be resistant? Does a phage pay a cost to infect resistant bacteria? Such tradeoffs may have interesting consequences for the evolutionary dynamics as well as diversity of this host-pathogen community.
Lab Overview
In this lab, your group will be given a bacterial host (H0) and a phage pathogen (P0). You will select for a host resistant to the pathogen (H1), and a "host-range mutant" pathogen (P1) that can infect the newly-resistant H1. You will then compete the original host (H0) against its resistant descendant (H1), as well as the original pathogen (P0) against its host-range mutant (P1).
Lab Logistics
This lab runs for slightly more than a week. Starting on Day 1 (Tuesday) of the Luria-Delbrück lab, you will plate out P0 and H0 on an LB agar dish to generate H1 mutants. On Day 2 (Wednesday), one member of each group will streak out a single colony from your Day 1 dish to ensure that the ancestral phage is removed from the bacteria. This should take about fifteen minutes. Day 3 (Thursday) requires one or two people from each group to create a liquid culture of H1 (this should also take approximately fifteen minutes). On Day 4 (Friday) one member of your group will plate ancestral phage (P0) in the presence of resistant bacteria (H1) in order to isolate host-range phage mutants (P1), which will not take more than half an hour. A TA will transfer these plates into the refrigerator on Saturday, and on Day 5 (Monday) the host-range mutant will be isolated, and both it and P0 must be titered. This will take one or two group members about 45 minutes. Day 6 (Tuesday) initiates both the pathogen competition and the bacteria competition, and on Day 7 (Wednesday) you will count the previous day's results and titer the end of both competitions (expect to have two group members spend about an hour on this). Day 8 (Thursday) only requires one person to count the results of this second titer, and should take no more than fifteen minutes.
Protocol
Day 1 (Tuesday)
Daily Overview
Plate ancestral bacteria (H0) in the presence of ancestral phage (P0) in order to isolate resistant bacteria (H1).
Materials
- 18mm tube containing P0
- 18mm tube containing H0
- 18mm tube containing sterile glass beads
- 18mm tube rack
- 1 LB Petri dish
- Pipettemen & tips
- Incubator (set to 37°C)
- Burner & Striker
- 70% ethanol spray bottle and Kimwipes
- Beaker with 95% ethanol for used glass beads
- Waste Bucket
- Gloves
Protocol
- Put on your gloves Ethanol your bench and pipette using a Kimwipe.
- Label the LB dish: "P0+H0, Day 1, today's date , your group's name"
- Label the tubes containing P0 and H0 with "today's date, your group's name"
- After the ethanol on your bench evaporates, turn on your gas and light your burner.
- Pour 6-10 sterile glass beads onto your labeled dish near the flame.
- Using a sterile tip, pipette 200µL of P0 onto the center of the LB dish. Discard tip into waste.
- Using a new sterile tip, carefully pipette 100µL of H0 onto the center of the LB dish. The bacteria (H0) should be deposited directly into the pool of phage (P0). Discard tip into waste.
- Gently shake the LB dish for approximately 20 seconds so that the glass beads evenly distribute the liquid across the surface of the LB dish (this is best accomplished by shaking back-and-forth and rotating the dish periodically). Wait at least 10 seconds, then flip the dish lid-side-down to collect the beads in the dish lid, and discard beads into the beaker with 95% ethanol.
- TURN OFF YOUR GAS
- Place Petri Dish in 37°C incubator lid-side-down (this will be incubated for ~24 hours).
- Place the labeled tubes containing P0 and H0 into the refrigerator.
- Clean up your bench and double check that your gas is turned OFF.
Day 2 (Wednesday)
Daily overview
Streak a colony of resistant bacteria (H1) onto LB dish with a wire loop to ensure that the ancestral phage is removed from the bacteria.
Materials
- LB dish with grown colonies (from Day 1, in the incubator)
- Wire loop
- One LB Petri dish
- Burner & Striker
- 70% ethanol spray bottle and Kimwipes
- Incubator (set to 37°C)
- Waste Bucket
- Gloves
Protocol
- Remove yesterday's LB dish from incubator and check to make sure there are colonies.
- Ethanol your bench.
- Label the new LB Petri dish: "H1 streak, Day 2, today's date, your group's name"
- After the ethanol evaporates, turn on your gas and light your burner.
- Away from the flame, wet Kimwipe with 70% ethanol and wipe your wire loop.
- Wait 10 seconds for ethanol to evaporate, then flame the loop and one inch of the wire until it is bright red. Cool the loop by waiting 10 seconds and then pressing somewhere on the agar in the new Petri dish (you should hear a little "sizzle").
- Using the wire loop, scoop up a single colony and streak onto the fresh, labeled LB dish.
- After completion of the streaking, flame the wire loop to sterilize it.
- Away from the flame, wet Kimwipe with 70% ethanol and wipe your wire loop.
- TURN OFF YOUR GAS
- Place freshly streaked Petri dish in 37°C incubator lid-side-down (this will be incubated for 24 hours).
- Clean up your bench and double check that your gas is turned OFF.
Day 3 (Thursday)
Daily Overview
Transfer isolated colony of resistant bacteria (H1) to liquid medium, initiating a growing, liquid culture.
Materials
- LB dish with streaked colonies (from Day 2, in the incubator)
- 18mm tube with 5 mL of LB Broth
- 18mm tube rack
- Wire loop
- Burner & Striker
- 70% ethanol spray bottle and Kimwipes
- Shaking incubator (set to 37°C)
- Waste Bucket
- Gloves
Protocol
- Retrieve Day 2 streaked LB dish from 37°'C incubator.
- Label 18 mm tube: "H1, Day 3, today's date, your group's name".
- Ethanol your bench.
- After the ethanol evaporates, turn on gas and light the burner.
- Clean the wire loop with ethanol, and wait 10 seconds until ethanol evaporates.
- Flame loop until it turns red. Cool the loop by pressing somewhere on the agar in the streaked Petri dish where there is no bacteria (you should hear a little "sizzle").
- Isolate a single colony from the streaked Petri dish with the wire loop.
- Insert the wire loop into labeled 18 mm tube containing LB broth and gently mix.
- Flame the wire loop to clean it.
- Clean the wire loop with ethanol.
- TURN OFF YOUR GAS.
- Insert 18mm tube into the shaking 37°C incubator (this will be incubated for 24 hours).
- Clean up your bench and double check that your gas is turned OFF.
Day 4 (Friday)
Daily Overview
Plate ancestral phage (P0) in the presence of resistant bacteria (H1) in order to isolate host-range phage mutants (P1).
Materials
- 18mm tube with 5 mL of H1 culture (from Day 3, in the incubator)
- 18mm tube containing P0 (in the refrigerator)
- 18mm tube rack
- 2 LB Petri dishes
- Dry bath with heat blocks (for 13mm tubes)
- 2 13mm tube with 4mL of soft agar held at 55°C
- 13mm tube rack
- Burner & Striker
- 70% ethanol spray bottle and Kimwipes
- Pipettemen and sterile tips
- Waste Bucket
- Gloves
Protocol
- Label 2 fresh LB dishes: "H1+P0, Day 4, today's date, your group's name"
- Retrieve the tube containing your H1 culture from the shaking 37°C incubator and the tube containing your P0 from the refrigerator.
- Ethanol your bench.
- After the ethanol evaporates, turn on gas and light burner
- What follows must occur in less than 1 minute, to prevent premature soft agar solidification.
- Pipette 400µL from the tube containing the H1 culture to the 13mm tube with soft agar and discard tip into waste.
- Pipette 100µL from the tube containing P0 into the same 13mm tube.
- Pour the contents of the soft agar tube onto the labeled LB dish and immediately (gently) swirl the dish until the soft agar uniformly covers the surface (this should all take only seconds).
- Put your now empty 13mm tube into the waste tube rack.
- Repeat previous step for the other LB dish.
- Let the dishes sit for 5 minutes without moving it (this will allow the soft agar to solidify).
- TURN OFF YOUR GAS
- Place your LB dishes in 37°C incubator lid-side-down (this will be incubated for ~24 hours and then placed in the refrigerator by your TA for the rest of the weekend).
- Put the tubes with H1 and P0 in the refrigerator.
- Clean up your bench and double check that your gas is turned OFF.
Day 5 (Monday)
Daily overview
Isolate the host-range phage mutant (P1) from Day 4 dish and titer both the ancestral (P0) and (P1) phage.
Materials
- 2 LB dishes with phage plaques (from Day 4, in the refrigerator)
- Culture of H0 (in the refrigerator)
- Culture of H1 (in the refrigerator)
- Tube containing P0 (in the refrigerator)
- 2 18mm tubes with 5mL LB
- 18mm tube rack
- Eppendorf tube with 50µL CHCl3 + 500µL saline
- Eppendorf tube rack
- Microtiter plate (with 270µL of saline in relevant wells)
- Dry bath with heat blocks (for 13mm tubes)
- 4 13mm tubes with 4mL of soft agar each (held at 55°C)
- 13mm tube rack
- 4 LB Petri dishes
- Burner & Striker
- 70% ethanol spray bottle and Kimwipes
- Pipettemen and tips
- Cut tips
- Waste Bucket
- Vortexer
- Gloves
Protocol
- Label Eppendorf tube: "P1, Day 5, today's date, your group's name"
- Retrieve the tubes containing your P0, H0, and H1 cultures as well as your LB dishes with phage plaques (from Day 4) from the refrigerator.
- Ethanol your bench.
- After the ethanol evaporates, turn on gas and light burner.
- From a dish containing P1 plaques labeled "H1 + P0", select a large plaque to isolate.
- Using a sterile cut pipette tip, take a plug from the center of the plaque. Then, dispense plug into labeled Eppendorf tube with chloroform and saline. (This can be done by setting the pipette to some low volume [e.g., 30µL] loading the cut tip, pressing the tip through the agar in the middle of a plaque, and aspirating; to dispense the plug into the Eppendorf tube, simply purge the pipette [if the plug is stuck in the tip, then washing with saline by aspirating and dispensing can sometimes liberate it]).
- After making sure the plug is in the Eppendorf tube, dispense cut tip into waste.
- Close the Eppendorf cap and vortex tube for approximately 30 seconds and let Eppendorf tube sit in the Eppendorf tube rack for at least 1 minute to allow chloroform to settle at bottom.
- While waiting, label your microtiter plate with your group's name. Then label the first column of your microtiter plate "P0" and the second column "P1".
- Using a sterile tip, pipette 30µL of your P0 phage from the tube you retrieved from the fridge, dispense to well A1 of the microtiter plate, and dispose of tip into waste.
- Using a new sterile tip, pipette 30µL of your P1 phage from the Eppendorf tube, dispense to well A2 of the microtiter plate, be careful to aspirate from the top layer (the bottom layer is chloroform) and dispose of tip into waste.
- Perform a dilution series to 10-6 (switching tips between each dilution) for both the P0 and the P1 phage.
- Label the four LB dishes:
- "P0 10-5, Day5, today's date, your group's name"
- "P0 10-6, Day5, today's date, your group's name"
- "P1 10-5, Day5, today's date, your group's name"
- "P1 10-6, Day5, today's date, your group's name"
- Vortex the H0 culture tube for 10 seconds.
- Using a sterile tip, pipette 400µL of H0 into one of the tubes with soft agar.
- Using a sterile tip, pipette 100µL of P0 from the 10-5 dilution (i.e., well E1) into the same tube with soft agar.
- After gentle mixing, pour the contents of the soft agar tube onto the LB dish labeled "P0 10-5" and immediately (gently) swirl the dish until the soft agar uniformly covers the surface (this should all take only seconds).
- Put your empty 13mm tube into the waste tube rack.
- Let the dish sit for 5 minutes without moving it (this will allow the soft agar to solidify).
- Repeat steps 15-19 replacing the phage and dilution in step 16 (P0 10-6, P1 10-5, and P1 10-6) and the appropriately labeled dish.
- Label two 18mm tubes:
- "H0, Day 5, today's date, your group's initials"
- "H1, Day 5, today's date, your group's initials"
- Using a sterile pipette tip, transfer 50µL from the vortexed H0 culture into the fresh tube labeled "H0, Day 5".
- Vortex the H1 culture tube (which you retrieved from the fridge) for 10 seconds. Using a sterile pipette tip, transfer 50µL from the vortexed H1 culture into the fresh tube labeled "H1, Day 5".
- TURN OFF YOUR GAS.
- Place your dishes in the 37°C incubator lid-side-down (this will be incubated for ~24 hours).
- Place the fresh 18mm tubes with bacterial culture into the 37°C shaking incubator (this will be incubated for ~24 hours).
- Place your microtiter plate, and P0 and P1 phage tubes and the old H0 and H1 bacteria tubes into the refrigerator.
- Clean up your bench and double check that your gas is turned OFF.
Day 6 (Tuesday)
Daily Overview
Initiate a competition between ancestral bacteria (H0) and resistant bacteria (H1) in the absence of phage. Initiate a competition between ancestral phage (P0) and host-range mutant phage (P1) competing for ancestral bacteria (H0).
Materials
- 4 LB dishes with phage plaques (from Day 5, in the incubator)
- Culture of H0 (from Day 5, in the shaking incubator)
- Culture of H1 (from Day 5, in the shaking incubator)
- Tube containing P0 (in the refrigerator)
- 2 18mm tubes with 5mL LB
- 18mm tube rack
- Eppendorf tube containing P1 (in the refrigerator)
- Eppendorf tube rack
- Microtiter plate (with 270µL of saline in relevant wells, in the refrigerator)
- Dry bath with heat blocks (for 13mm tubes)
- 4 13mm tubes with 4mL of soft agar each (held at 55°C)
- 13mm tube rack
- 8 LB Petri dishes
- Burner & Striker
- 70% ethanol spray bottle and Kimwipes
- Pipettemen and tips
- Waste Bucket
- Vortexer
- Sterile glass beads
- Beaker with 95% ethanol for glass beads
- Clicker Counter and marker
- Gloves
Protocol
- Retrieve all dishes from the incubator.
- Count the phage plaques from the dishes incubated on Day 5.
- Calculate the titer of phage in your tubes in the refrigerator (see Calculating Titers? protocol for details).
- Use the power of math to calculate the dilution and volume of phage you need in order to start a competition where both P0 and P1 start at a titer of 5 x 105 PFU/mL in a tube containing 5mL of LB.
- Check your work with your TA.
- Retrieve your tubes of P0 and P1 and the microtiter plate from the fridge as well as the H0 and H1 cultures from the shaking incubator.
- Label one 18mm tube: "Phage Comp, Day 6, today's date, your group's name"
- Using your calculated phage dilutions and volumes, add calculated volumes of P0 and P1 (re-dilute in new saline wells, and be careful to avoid chloroform at the bottom of some tubes) to the 18mm tube labeled "Phage Comp, Day 6" and vortex tube for 30 seconds.
- Label column 4 of the microtiter plate "Phage. Comp".
- Using a sterile tip, pipette 30µL from the "Phage Comp" tube to the fourth column of the microtiter plate (well A4).
- Perform a dilution series to 10-3 (switching tips between each dilution) for this phage mixture.
- Label four LB dishes (although we always strive to be politically correct, PC stands for "Phage Competition" below):
- "PC H0 10-2, Day 6, today's date, your group's name"
- "PC H0 10-3, Day 6, today's date, your group's name"
- "PC H1 10-2,, Day 6, today's date, your group's name"
- "PC H1 10-3, Day 6, today's date, your group's name"
- Titer the Phage Competition:
- Using a sterile tip, pipette 400µL of H0 into one of the tubes with soft agar.
- Using a sterile tip, pipette 100µL of phage mixture from the 10-2 dilution (i.e., well B4) into the same soft agar tube.
- After gentle mixing, pour the contents of the soft agar tube onto the LB dish labeled "PC H0 10-2" and immediately (gently) swirl the dish until the soft agar uniformly covers the surface (this should all take only seconds).
- Put your empty 13mm tube into the waste tube rack.
- Let the dish sit for 5 minutes without moving it (this will allow the soft agar to solidify).
- Repeat previous step replacing the bacteria used (H0, H1, and H1), the dilution (10-3, 10-2, 10-3) and the appropriately labeled dish.
- Add 500µL of H0 to "Phage Comp." tube (this initiates the phage competition).
- Place the "Phage Comp." tube into the 37°C shaking incubator (this will incubate for ~24 hours).
- Label the second 18mm tube: "Host Comp, Day 6, today's date, your group's name".
- Label column 6 of the microtiter plate "Host Comp".
- Using a sterile tip, pipette 50µL of H0 from fully grown culture into the tube labeled "Host Comp, Day 6" and dispose of tip into waste. Using a sterile tip, pipette 50µL of H1 from fully grown culture into the "Host Comp, Day 6" tube and dispose of tip into waste. Vortex the competition tube for 30 seconds.
- Using a sterile tip, pipette 30µL from the host competition to the sixth column of the microtiter plate (well A6).
- Perform a dilution series to 10-5 (switching tips between each dilution) for this bacteria mixture.
- Label four LB dishes (HC stands for "Host Competition"):
- "HC P0 10-4, Day 6, today's date, your group's name"
- "HC P0 10-5, Day 6, today's date, your group's name"
- "HC blank 10-4 Day 6, today's date, your group's name"
- "HC blank 10-5, Day 6, today's date, your group's name"
- Titer host competition with P0:
- Pour 6-10 sterile glass beads onto each of your labeled dishes near the flame.
- Using a sterile tip, pipette 200µL of P0 onto the center of the LB dish labeled "HC P0 10-4". Discard tip into waste.
- Using a new sterile tip, carefully pipette 100µL of the 10-4 dilution of the bacterial mixture onto the center of the dish. The bacteria should be deposited directly into the pool of phage (P0). Discard tip into waste.
- Gently shake the Petri dish for approximately 20 seconds so that the glass beads evenly distribute the liquid across the surface of the Petri dish and then discard beads into beaker with 95% ethanol.
- Repeat previous step replacing the dish ("HC P0 10-5") and the dilution (10-5).
- Titer host competition without P0:
- Using a sterile tip, pipette 100µL of the 10-4 dilution of the bacterial mixture (well B6) onto the center of the dish labeled "HC blank 10-4". Discard tip into waste. There should be no phage on this plate (which is what "blank" indicates).
- Shake the Petri dish and then discard beads into beaker with 95% ethanol.
- Repeat previous step replacing the dish ("HC blank 10-5") and the dilution (10-5).
- TURN OFF YOUR GAS
- Place the bacteria competition tube into the 37°C shaking incubator and the dishes in the 37°C incubator (these will all incubate for ~24 hours).
- Place the P0, P1, H0 and H1 cultures and your microtiter plate, in the refrigerator.
- Clean up your bench and double check that your gas is turned OFF.
Day 7 (Wednesday)
Daily Overview
Titer the end of the host (H0 versus H1) and pathogen (P0 versus P1) competition, and count the previous day's results.
Materials
- 8 LB dishes with phage plaques or bacterial colonies (from Day 6, in the incubator)
- "Host Competition" tube (from Day 6, in the shaking incubator)
- "Phage Competition" tube (from Day 6, in the shaking incubator)
- Culture of H0 (from Day 5, in the refrigerator)
- Culture of H1 (from Day 5, in the refrigerator)
- One tube containing P0 (in the refrigerator)
- 18mm tube rack
- Microtiter plate (with 270µL of saline in relevant wells, in the refrigerator)
- Dry bath with heat blocks (for 13mm tubes)
- 6 13mm tubes with 4mL of soft agar each (held at 550C)
- 13mm tube rack
- 12 LB Petri dishes
- Burner & Striker
- 70% ethanol spray bottle and Kimwipes
- Pipettemen and tips
- Waste Bucket
- Sterile glass beads
- Beaker with 95% ethanol for glass beads
- Clicker counter and marker
- Gloves
Protocol
- Retrieve all dishes from the incubator.
- Retrieve your phage competition tube (from the shaking incubator), the microtiter plate, and H0 and H1 day 5 cultures from the refrigerator.
- Label column 7 of the microtiter plate "Phage Comp End".
- Using a sterile tip, pipette 30µL from the "Phage Comp" tube to the seventh column of the microtiter plate (well A7).
- Perform a dilution series to 10-7 (switching tips between each dilution) for this phage mixture.
- Label six LB dishes (PC stands for "Pathogen Competition" below):
- "PC H0 10-5, Day 7, today's date, your group's name"
- "PC H0 10-6, Day 7, today's date, your group's name"
- "PC H0 10-7, Day 7, today's date, your group's name"
- "PC H1 10-5, Day 7, today's date, your group's name"
- "PC H1 10-6, Day 7, today's date, your group's name"
- "PC H1 10-7, Day 7, today's date, your group's name"
- Titer the phage competition
- Using a sterile tip, pipette 400µL of H0 into one of the tubes with soft agar.
- Using a sterile tip, pipette 100µL of phage mixture from the 10-5 dilution (i.e., well E7) into the same soft agar tube.
- Pour the contents of the soft agar tube onto the LB dish labeled "PC H0 10-5" and immediately (gently) swirl the dish until the soft agar uniformly covers the surface (this should all take only seconds).
- Put your empty 13mm tube into the waste tube rack.
- Let the dish sit for 5 minutes without moving it (this will allow the soft agar to solidify).
- Repeat previous step replacing the bacteria (H0 or H1), the dilution (10-5, 10-6 and 10-7) and the respective dish.
- Place the "Phage Comp.", H0 and H1 tubes into refrigerator.
- Label column 8 of the microtiter plate "Host Comp End".
- Using a sterile tip, pipette 30µL from the host competition to the eighth column of the microtiter plate.
- Perform a dilution series to 10-7 (switching tips between each dilution) for this bacteria mixture.
- Label six LB dishes (HC stands for "Host Competition"):
- "HC P0 10-5, Day 7, today's date, your group's name"
- "HC P0 10-6, Day 7, today's date, your group's name"
- "HC P0 10-7, Day 7, today's date, your group's name"
- "HC blank 10-5, Day 7, today's date, your group's name"
- "HC blank 10-6, Day 7, today's date, your group's name"
- "HC blank 10-7, Day 7, today's date, your group's name"
- Titer Host Competition with P0.
- Pour 6-10 sterile glass beads onto each of your labeled dishes near the flame.
- Using a sterile tip, pipette 200µL of P0 onto the center of the LB dish labeled "HC P0 10-5". Discard tip into waste.
- Using a new sterile tip, carefully pipette 100µL of the 10-5 dilution of the bacterial mixture onto the center of the dish. The bacteria should be deposited directly into the pool of phage (P0). Discard tip into waste.
- Gently shake the Petri dish for approximately 20 seconds so that the glass beads evenly distribute the liquid across the surface of the Petri dish and then discard beads into beaker with 95% ethanol.
- Repeat previous step replacing the dish ("HC P0 10-6 and HC P0 10-7") and the dilution (10-6 and 10-7).
- Titer Host Competition without P0.
- Using a sterile tip, pipette 100µL of the 10-5 dilution of the bacterial mixture onto the center of the dish labeled "HC blank 105". Discard tip into waste. There should be no phage on this plate (which is what "blank" indicates).
- Shake the Petri dish and then discard beads into beaker with 95% ethanol.
- Repeat previous step replacing the dish ("HC blank 10-6" and "HC blank 10-7") and the dilution (10-6 and 10-7).
- Place the "Host Comp." and P0 tubes into refrigerator.
- TURN OFF YOUR GAS
- Place your LB dishes in 37°C incubator lid-side-down (they will be incubated for ~24 hours).
- Count yesterday's dishes. Record the data on the dishes, in your lab notebook, and e-mail it to any lab members that were not able to attend today. Make sure to enter your data in the class spreadsheet.
Day 8 (Thursday)
Daily Overview
Conclude the Evolutionary Arms Race lab by counting the final pathogen and host competition dishes.
Materials
- 12 LB dishes with phage plaques or bacterial colonies (from Day 7, in the incubator)
- Clicker counter and marker
- Gloves
Protocol
- Count yesterday's dishes. Record the data on the dishes, in your lab notebook, and e-mail it to any lab members that were not able to attend today. Make sure to enter your data in the class spreadsheet.
Putting it all together
After finishing your counting of all the dishes with colonies and plaques, you should be able to calculate the abundance of each competitor (X0 or X1, where “X” is either “P” or “H”) at the beginning and end of each competition (phage or host).
Note that in each case, the count on one dish will allow you to compute the concentration of X0+X1 and the count on another dish will allow you to compute the concentration X1. For instance, counting plaques on a plate where H0 forms the lawn will allow you to calculate the concentration of P0+P1 (since both the ancestor and host-range mutant form plaques on H0); whereas, counting plaques on a dish where H1 forms the lawn will allow you to compute the concentration of P1 (since only the host-range mutant forms plaques on H1). Thus, in order to calculate the concentration of X0, you will have to subtract one estimate (concentration of X1) from the other (pooled concentration of X0+X1).
Let the concentration of X0 and X1 (cells or phage) at the beginning of the assay be given by x0,b and x1,b, respectively, and the number of ancestor and evolved cells at the end of the experiment be given by x0,e and x1,e, respectively. The fitness of X1 relative to X0 (its ancestor) is given by:
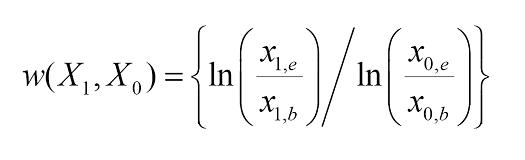
Using this equation, you should be able to compute the fitness of the resistant host relative to its sensitive ancestor as well as the fitness of the host-range phage mutant relative to its ancestor. In particular, you should be able to assess whether there are costs to counter-adaptations in this arms race.
Questions
- Report the fitness of your phage-resistant bacterial host relative to its sensitive ancestor and the fitness of your host-range mutant phage relative to its ancestor.
- Do you have evidence for costs of counter-adaptation in this victim-exploiter interaction? Using the class data, what can you say statistically about these costs? (Provide your reasoning for the test you employ.)
- Discuss the P0 and P1 phage in terms of the specialist-generalist distinction. Do you have evidence that being of jack-of-all-trades means that you can master none?
- Consider a community where resistant bacteria and host-range phage readily arise by mutation, but involve costs relative to their ancestors. Do you think such costs will play positive or negative roles in promoting diversity in this community? Explain your reasoning.
- Suppose you plate a dilution from your P1 tube in soft agar with the H0 host and then separately again with the H1 host. You find that the number of plaques on the H1 host is some fraction of the number of plaques on the H0 host. Do you expect this? If so, explain why. If not, provide a hypothesis about what might account for this discrepancy.
- Bonus Question: If H1 is plated with P1, then sometimes very small colonies will appear. Let’s call these bacteria H2. How would you experimentally assess the fitness of H2 relative to H1? How would experimentally assess the fitness of H2 relative to H0? (If you plan to use competitions, give details of how you will plate the competitors). What do you imagine would be likely dynamics for the community under the following scenarios:
- w(H2,H0) < w(H2,H1) < 1
- w(H2,H0) = w(H2,H1) = 1