Bio 482 POP
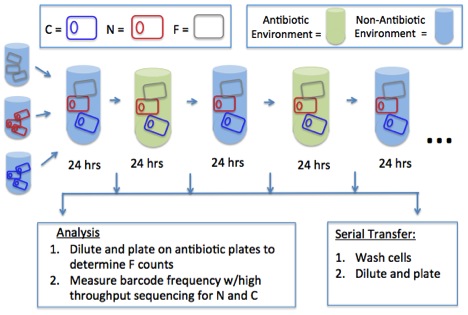
Fig: A schematic of my experimental set-up
Research Description
Unlike eukaryotes, most prokaryotes exchange genes horizontally and can do so at a high frequency. Plasmid transfer via conjugation is the most common mechanism of horizontal gene transfer in bacteria. It is still unclear, however, how conjugative plasmids persist in populations given the costs associated with conjugation. Furthermore, given such costs, why are some genes found on conjugative plasmids rather than the chromosome?
My preliminary results via mathematical modeling indicate that conjugative plasmids can be maintained in an environment with alternating selection. I will test this hypothesis experimentally using a biological system consisting of three players: an E. coli strain containing a conjugation-proficient plasmid, a conjugation-deficient plasmid, and a plasmid-free strain. I will compete these three strains in an environment that a) constantly selects for the plasmid-encoded gene (antibiotic present), b) constantly selects against the plasmid-encoded gene (antibiotic-free), and c) alternates with periodic switching between the antibiotic-present and antibiotic-free environments. This will involve estimating the parameters of my experimental system (e.g. conjugation rates, segregation rates, plasmid carraige costs, etc.) and monitoring these parameters throughout my proposed experiment to determine any evolutionary changes that may occur.
This project will provide new insights into how and why conjugative plasmids persist. Given the critical role that plasmids play in the spread of antibiotic resistance genes, this work also has wider implications for human health by contributing to our understanding of the impact of antibiotic cycling on antibiotic resistance.
Back to Biology 482 Homepage